January 28, 2016 — Back in 2011, Jennifer Doudna, a biochemist and molecular biologist at the University of California, Berkeley, and Emmanuelle Charpentier, now at the Max Planck Institute for Infection Biology in Germany, grew intrigued by the way bacteria use a molecular system known as CRISPR-Cas9 to respond to viral attacks. For years, bacteria were assumed to be primitive creatures with rudimentary immune systems. But CRISPR-Cas9 revealed a startlingly sophisticated memory-response scheme. The bacteria store DNA samples from invading viruses by tucking them into a DNA library called CRISPR that is part of the bacteria’s natural genome. If the same virus should attack again, the Cas9 enzyme is primed by the CRISPR library to cut (and thus disable) viral DNA with the same sequence.
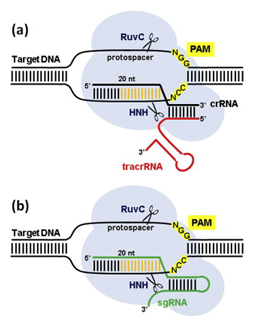
In the native bacterial system (a), a structure that’s formed by crRNA and tracrRNA and includes a “guide” segment (gold) guides the Cas9 protein (light blue blob) to a spot in the viral DNA that corresponds to the guide segment. The cRNA is critical in targeting while tracRNA stabilizes the structure and activates Cas9 to cleave the DNA. To turn this natural system into a useful tool for genetic manipulation (b), researchers created an artificial single guide RNA molecule (sgRNA, in green) by fusing the crRNA and tracrRNA. Image courtesy of Elsevier
After months of trying to tease apart how the system works, Doudna’s team determined that two RNA molecules play central roles: CRISPR RNA (crRNA), which leads Cas9 to a particular location on the viral gene, and a trans-activating RNA (tracrRNA), which helps activate Cas9. Together, these two RNA molecules empower Cas9 to make its cuts.
Still, it was not clear that CRISPR would be all that exciting or useful outside of bacteria. Microbes have very different cell structures than animals and plants, and it was quite possible that the system would only work in bacteria. The real breakthrough occurred in 2012 when Doudna, Charpentier and then-postdoctoral fellow Martin Jinek realized it would be possible to combine the crRNA and the tracrRNA into a single, artificial guide RNA (sgRNA). By adding to the sgRNA a customized “guide segment” matching a particular DNA sequence in an organism of interest, they could aim Cas9 to cut any organism’s genome in any spot they wished.
Often likened to a word processor, CRISPR can be used to target whole gene “words” or a few nucleotide “letters” with precision and speed that far outpaces conventional genetic engineering. It’s a superb tool for deleting chunks of DNA and for facilitating precise substitutions when researchers want to swap a few key nucleotide sequences.
Less often emphasized is that CRISPR can also be used to add new genes or parts thereof. The key here is understanding what happens after Cas9 makes its cuts.
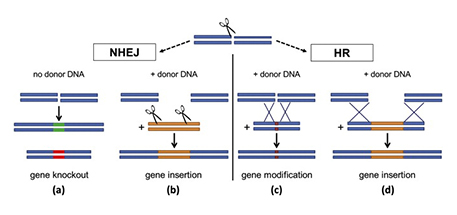
A Cas9-caused break in DNA can be repaired in four different ways, two of which open the door to inserting a new gene of choice. Image courtesy of Elsevier
The cell’s DNA repair machinery typically takes over in one of two different modes. In the first mode (called “non-homologous end joining,” or NHEJ), it usually glues the two pieces back together, but imperfectly, deactivating the gene (see “a” above). Such “gene knockouts” don’t involve any foreign DNA but can eliminate traits that affect food quality, confer susceptibility to diseases or divert energy away from valuable end products such as grain or fruit. Occasionally, say researchers, this pathway may leave a DNA cut with “sticky ends,” enabling foreign genes of interest to be directly spliced in (b) — a double-stranded DNA insertion somewhat akin to “old-fashioned” genetic engineering.
A second kind of repair (called “homology-directed repair” or “homologous recombination” — HR) is much less common but far more accurate. In HR, the cut ends aren’t just jammed back together; the cell machinery copies a nearby piece of DNA to fix the damaged sequence. By providing a DNA snippet of their choice, scientists can induce the cell to fill in any desired sequence, from a small mutation (c) to a whole new gene (d). This HR pathway, says Fuguo Jiang, a postdoctoral fellow in Doudna’s lab, is not yet fully understood. But, as this illustration shows, it involves a meticulous process of one strand of donor DNA being stitched into the host gene, providing the template for cellular repair. ![]()
Editor’s note: For more on CRISPR, please see “CRISPR is coming to agriculture — with big implications for food, farmers, consumers and nature.”
Ensia shares solutions-focused stories free of charge through our online magazine and partner media. That means audiences around the world have ready access to stories that can — and do — help them shape a better future. If you value our work, please show your support today.
Yes, I'll support Ensia!
