August 4, 2014 — For years, Monterey Bay Aquarium’s 1.2 million gallon main tank has been a source of thrill for visitors seeking fish-eye views of life in the pounding Pacific Ocean right outside its doors. Now, thanks to advances in DNA sequencing, it’s also becoming a model for how to survey life in that vast ocean with just a few glasses of water.
In a study published last January in PLoS ONE, researchers led by Ryan Kelly of the University of Washington identified most of the bony fishes in the aquarium’s great tank by deciphering the DNA in samples of the tank’s water.
Environmental DNA sampling is the latest tool in biodiversity monitoring in marine and aquatic systems. Advanced technologies now make it possible to associate bits of DNA found in the environment with genetic sequences found in various species. Scientists have used eDNA sampling to detect the former presence of ancient plants and animals on land since at least 2003, but only recently have they begun to test its accuracy in water.
The ability to assess wild fish stocks is becoming increasingly important as commercial fisheries collapse worldwide.
Results from studies in freshwater ecosystems are promising. Researchers have detected invasive sunfish in Japanese ponds and secretive amphibians in Rocky Mountain streams using eDNA.
Its potential in marine ecosystems is even more alluring. The ability to assess wild fish stocks is becoming increasingly important as commercial fisheries collapse worldwide, and coral reefs threatened by climate change, coastal pollution, overfishing and invasive species need careful monitoring to protect their diverse ecosystems. Yet traditional marine surveys typically require expensive boats, nets and other gear that cash-strapped researchers might not have.
Get More, Pay Less
“Ship time alone is $50,000 a day,” says Kelly. “Then, if you’re paying divers to survey a coral reef, for example, you’re paying several hundred dollars an hour plus expenses. [Yet] divers are going to focus on [only] a handful of species. … How many thousands of species of worms live in a coral reef that don’t get counted by the diver? But they’re all giving off DNA.”
Kelly and other scientists note that traditional marine and freshwater survey techniques are rarely standardized, making it difficult to compare data across studies and time. And they ultimately rely on taxonomic expertise to identify the creatures counted.
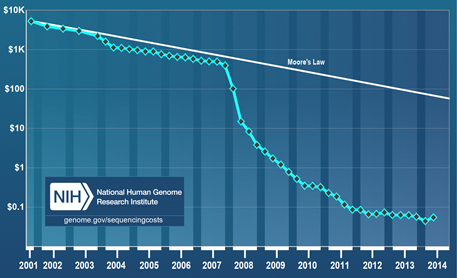
Huge declines in the cost per raw megabase of DNA sequence in recent years have made environmental DNA analysis a viable option for deducing the presence of species from water samples. Image courtesy of the National Human Genome Research Institute.
In contrast, the cost of gene sequencing has plummeted in recent years. Processes that cost $5,300 in 2001 cost only about 5 cents today. For the Monterey Bay study, that means that Kelly and his colleagues could get about 10 million reads of the gene segment that interested them for about $1,300. With that, they identified four of the five bony fishes in the tank down to the family or genus level from only a handful of water samples. Not only that, but the differences in the amount of the various types of DNA found in the samples accurately reflected the known composition of the tank, with sardines and tuna showing up as the most abundant species and dolphinfish and chub mackerel as the least.
The technique isn’t perfect, however: The study failed to detect the tank’s sharks, rays and sea turtle. Kelly attributes that to the genetic primers his team used. Primers are short segments of DNA that bind with identical segments in samples. Once they latch on, the sample segments replicate themselves. The replicates are then used to identify each species. Kelly believes the set of primers he used didn’t include genetic material found in sharks, rays or turtles.
Promising Tool
How sensitive is the approach? In 2012, Philip Francis Thomsen and Eske Willerslev of the University of Copenhagen tested eDNA techniques in wild freshwater and marine ecosystems in Europe. At 98 freshwater locations they and their co-authors targeted six rare animals known to inhabit those sites, and detected each in most or all of their samples. Moreover, using controlled experiments in artificial ponds, they confirmed that when more animals were present, the concentration of their DNA increased. At three coastal sites in Denmark, they identified 15 species of fish — far surpassing the detection levels of traditional survey methods — and several rare animals, including the red-throated loon, an occasional seasonal migrant. “We’ve found the DNA even from mammals that were just drinking the water and left some DNA behind, so that says something about the sensitivity of this approach,” says Willerslev.
The extreme sensitivity of eDNA surveying makes it a promising tool for detecting species that traditional surveys miss, and invasive species before they become a problem. But measuring population sizes with eDNA remains elusive. DNA concentrations depend not only on how many animals are present, but the animals’ size, shape and rate at which they shed cells and waste containing DNA. Water temperature, salinity and currents can affect DNA degradation and dispersal, altering concentrations.

Ryan Kelly and graduate student Natalie Lowell take a sample in Seattle as part of a project comparing the eDNA “signatures” of freshwater and saltwater. Photo by Kathryn Graziano.
Alexandria Boehm, an associate professor of civil and environmental engineering at Stanford University, is studying DNA shedding and degradation rates as a step towards being able to use eDNA quantitatively. “As an engineer, I tend to think about sources, fluxes, transport and rates,” Boehm says. A better understanding of these relationships, she says, could help open the door to being able to use eDNA to estimate abundance as well as presence of species.
Ultimately, the usefulness of eDNA depends on genetic sequencing of Earth’s biodiversity. The National Institutes of Health database GenBank, which stores genetic information on species from around the world, has grown exponentially since its founding in 1982. But it doesn’t yet include everything.
Regardless, eDNA already is proving its worth. In fact, the U.S. Fish and Wildlife Service is using eDNA techniques to monitor whether invasive Asian carp have entered the Great Lakes and other vulnerable waterways. And Kelly is testing the technique with scientists from the National Marine Fisheries Service to replace manual surveys of animals living in eelgrass beds in Washington’s Puget Sound.
Ensia shares solutions-focused stories free of charge through our online magazine and partner media. That means audiences around the world have ready access to stories that can — and do — help them shape a better future. If you value our work, please show your support today.
Yes, I'll support Ensia!
